Our Research
Development of molecular and imaging methods and their applications in biology and biomedicine
At present, the genomes of many animals have been sequenced and thousands of mutant worms, flies, and mice have been produced. Despite this progress, we do not understand what the vast majority of genes and their proteins actually do in many organisms. Even for those proteins whose function has been identified, the precise nature of how the proteins function is not known since most functional conclusions about a protein deduced from molecular biology methods are indirect.
One of the major problems is that we cannot currently see what a protein does, where it is, and how it moves. The aim of our group is to develop a set of novel optical probes to add to the current suite of powerful optical and electron microscopy methods. The development of these new probes is driven by our desire to visualize the molecular organization and dynamics. Our ultimate goal is the visualization of proteins functioning in cellular processes at nanometer resolution in real time with molecular resolution.
Many applications of fluorescence microscopy benefit from nanometer-sized, bright, photostable, bio-specific, non-toxic probes that can be used to label biomolecules in live cells inside behaving animals and in tissue culture. These probes would be immediately applicable to a series of outstanding questions in cellular biology and biomedicine such as the investigation of cell signaling, cell development and differentiation, and cancer metastasis.
Therefore, we are developing a set of novel imaging tools to add to the current suite of microscopy methods. These include the development of photostable optical probes to label single proteins in cells, tissues and organisms. In thin biological samples, we are aiming for live cell imaging with ~10 nanometer spatial resolution. Our approach is based on the development of fluorescent rare earth ions imbedded in crystalline host lattices and silicon-vacancies (SiV) in nanodiamonds.
We are investigating a new approach to achieving super-resolution structural information using many different fluorescent probes. The development is based on the fact that these probes can be addressed and detected independently through either their narrow line-width excitation and/or emission properties. Since thousands of impurity ions are embedded in each nanoparticle, altering the relative ratio of the impurity ions allows the creation of many spectrally identifiable probes.
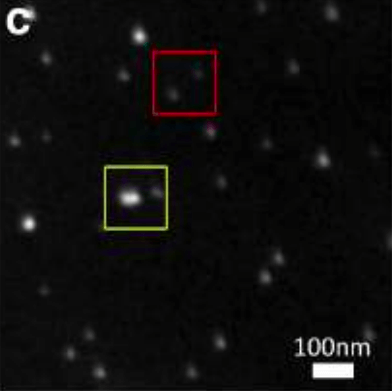
We have also developed methods to correlate the size and fluorescence brightness of ensembles of individual particles, which is essential for the full characterization of these fluorescent probes (Wisser, D., et al., ACS Photonics 3,1523 (2016)). With this characterization of the optical and physical morphology of our nanoparticles, we are further optimizing the brightness of particles with different dopant concentration, protective shell thickness and co-doping.
The biological applications include imaging multiple proteins optically using novel nanoprobes in fixed but hydrated samples of cells and tissue slices. We are also exploring novel applications in live cells and organisms where a combination of several nanoparticle probes with fluorescent proteins or cell-diffusible organic dyes can be used. The optical microscopy techniques will enable the imaging of multiple proteins tagged with rare-earth nanoparticles of different colors. These rare-earth nanoparticles (and color center nanodiamonds discussed below) are photostable and could potentially form the basis for Förster Resonance Energy Transfer (FRET) measurements. If successful, we would be able to observe protein-protein interactions over long time scales (minutes to hours) and cell development and migration over days. We believe that the nano-probes that we are developing may help solve many of the outstanding issues in fluorescence imaging. We outline the technological developments that constitute the major thrust of our lab, and discuss several biological questions we hope to address.
Synthesis and characterization of rare-earth nanoparticles
We are working with up-converting nanoparticles with Er3+ and Yb3+ imbedded in fluoride hosts. Up-converting nanoparticles have the advantage that excitation at wavelengths of 980 nm or 1064 nm are well-isolated from emission at ~545 nm for Er3+ and Yb3+ and 800 nm for Tm3+ and Yb3+ mixtures. We have made significant progress in properly characterizing rare-earth nanoparticle luminescence. In addition, the brightness of these particles has been increased substantially. These nanoprobes are becoming commercially available, and novel schemes to further increase the brightness continue to be invented. While these particles have comparable brightness to quantum dots, they are photostable, do not blink, and are significantly less chemically toxic. The background light from both biological auto-fluorescence and scattered light is greatly reduced due to the up-conversion from infrared light to visible or higher frequency process. We estimate that a few hundred counts (instead of a few thousand counts) per camera frame image are needed to obtain a good emission signal. The particles also allow multi-color spectral emission. We believe that roughly half a dozen different proteins can be imaged at the same time.
Nanoprobes with biochemical specificity for optical and electron microscopy
Another feature that differentiates rare earth nanoparticles from fluorescent proteins, organic dyes or quantum dots is that their luminescence properties withstand electron microscope irradiation. One of our goals is to image multiple proteins along with the cellular ultrastructure such as the plasma membrane, cellular organelles and vesicles in tissue samples using electron microscopy. Our approach is to use the cathodoluminescence properties of these nanoparticles: when the nanoparticles are bombarded with 1-30 keV electrons, they emit light. A biological cell labeled with these nanoparticles can be probed in an electron microscope with parallel electron and photon detection to retrieve both ultrastructural and biochemical information in a single experiment.
Just like in the case of electron microscopy, the interaction volume of electrons determines the spatial resolution in cathodoluminescence microscopy. Ultimately, we would like to use serial block face electron microscopy in combination with cathodoluminescence microscopy. This approach would allow tracing the connections inside the brain and imaging the biochemical nature of synapses in parallel.
Improved synthesis and characterization of nanodiamond probes
Exceptionally bright luminescence from SiV nanodiamonds at ~740 nm with count rates of ~ 5 million photons per second per SiV color center have been reported in the literature. In addition, there are several publications reporting the synthesis of diamond nanoparticles with less than 5 nm diameter. For this reason, we began to pursue the controlled synthesis of ultra-small nanodiamonds starting from molecular-size diamonds, “diamondoids”, originally found as a product of oil refining. This work is being done in collaboration with Professors Nick Melosh (Stanford Materials Science) and Z-X Shen (Stanford Applied Physics) who have been leaders in the growth of nanodiamonds from diamondoids.
We have developed a new CVD method of growing very high quality nanodiamonds, beginning with diamondoid seeds. A silicon dioxide layer, where the diamondoid seeds were tethered, turns out to be an effective source of silicon atoms for silicon vacancy formation. Photoluminescence measurements of these nanodiamonds at room temperature show a strong peak at ~740 nm with the characteristic weak phonon sideband. Using this method, chromium has also been incorporated into CVD diamond. The easy incorporation of chromium suggests it might be fairly straightforward to create a wide variety of desired color centers, e.g. the rare earth elements. In addition, with the growth time calibrated to the desired nanoparticle size, we are able to observe extremely high quality diamond lattices on small size nanodiamonds down to 10 nm.
Our CVD growth process can be modified to grow high quality nanodiamonds on resonator structures for quantum information and computation applications. For example, in a collaboration led by Prof. Jelena Vuckovic, we have used this process to grow nanodiamonds on silicon carbide-nanodiamond micro-resonators that enhance both SiV- and Cr-related color center via the Purcell enhancement.
Development of nanoparticle functionalization strategy
The utility of nanoparticle probes is critically dependent on the ability to passivate rare-earth nanoparticles nanodiamonds to minimize nonspecific binding to cells and also to label specific proteins of interest. The functionalized nanoparticles need to have high water-solubility and be fully biocompatible. In case of rare earth nanoparticles, their surface needs to be protected from water to prevent the quenching via energy coupling to the OH stretch vibration of water. In the case of NV, SiV, Cr and Ge color nanodiamonds, effective molecular labeling schemes need to be developed. In addition, it is vital to minimize the thickness of any surface coating that fulfills all these requirements.
For this purpose, we are exploring several surface functionalization strategies including the amphiphilic polymer wrapping strategies and few nanometer thick silica/PEG coatings that will allow functionalization of the NPs for targeted labeling with antibodies, nanobodies, SNAP-tag, and HaloTag ligands.
Nanoparticle cell tracking
One application of our nanoparticles is to deliver them into the cytosol and track cells through generations. The excellent photo stability of the nanoparticles we are investigating would allow us to do long term cell tracking. We have demonstrated that good mono-dispersity in cytosol will in general lead to roughly equal partitioning during cell division. With enough initial population of delivered nanoparticles, one may estimate cell generations before nanoparticles get diluted to single particle per cell level. We have begun collaborating with Stanford professors Kang Shen to study the development of C. elegans, Liqun Luo to investigate Drosophila, and Hiromitsu Nakauchi to study developing mouse embryos. We tested nanoparticle microinjection into mouse zygotes upon fertilization. In initial results, we found that the survival rate of fertilized eggs (to E13.5) injected with ~100,000 fluorescent nanodiamonds (FNDs) and tens of thousands of rare earth doped up-conversion nanoparticles (UCNP) and implanted into surrogate mouse mothers is comparable to nanoparticle-free cells. With proper functionalization coatings, UCNPs and luminescent nanodiamonds may allow us to track embryo development. As a first application, we want to locate FND containing cells in embryos when most organs begin to emerge. Cells containing multiple FNDs indicates a slow division speed and may possess the potential stem cell properties. However, these slow dividing cells could well be some cells that serve a special purpose such as hematopoietic stem cells (HSCs) and primordial germ cells (PGCs). We are currently designing experiments that will study how fluorescent nanodiamonds and rare earth nanoparticle donor cells contribute to chimera host development.
In addition, if our work in cathodoluminescence and EM imaging is successful, serial block-face EM imaging may allow us to identify the position of the nanoparticles with 20 nanometer precision in the context of developing embryos. Within mature organisms, cathodoluminescence and EM imaging may allow us to correlate the expression of specific proteins to changes in cell morphology such as in stages of synapse formation.
Adaptive optics imaging of nanoparticles
Obtaining optical resolution deep (>100 μm) inside living biological tissue presents unique opportunities for imaging rare-earth nanoparticles and studying protein interactions and cell development in vivo. However, index of refraction mismatches from different cells as well as scattering from thick tissue results in images that are severely blurred. To account for these deleterious effects, we've designed and assembled an adaptive optics system to provide real-time, widefield, image correction several hundreds of micrometers into biological samples.
The design of the adaptive optics system was heavily influenced by extensive literature reviews and state-of-the-art adaptive optics technology. The system is equipped with a Boston Micromachines Corporation’s microelectrical mechanical systems (MEMS) Kilo-DM to provide sample-induced aberration correction. A Princeton Instrument’s NIRvana 640 InGaAs detector gives us optimal coverage over the ideal biological deep tissue imaging range between 800 nm to 1.5 μm emission. The adaptive optics system is designed to operate without a wavefront sensor, and instead derives deformable mirror correction algorithms to converge on adaptive optics corrections. We are investigating a number of algorithms, including machine learning approaches to wavefront correction.
Acoustic imaging
Ultrasound imaging is an important medical diagnostics tool. It offers low cost, real-time imaging with no exposure to ionizing radiation. Refinements such as color Doppler, shear wave and contrast agent labeling offer valuable additional diagnostic information which complement x-ray, CT and MRI imaging modalities. However, ultrasound imaging is subject to significant speckle noise that severely degrades the effective resolution and limits its clinical use.
It is well-known that speckle can be reduced by compounding (averaging) images taken either at different frequencies or different angles. However, it has not been appreciated that a further reduction in speckle is achieved by combining frequency and angle compounding methods. We demonstrate that speckle is reduced multiplicatively due to the statistical independence of speckle variation with frequency and angle. Non-rigid image matching is applied to correct the distortions when the images are acquired from different angles and marked improvement in image quality is demonstrated.
Structural dynamics of G-proteins coupled receptor with photon stamped single molecule FRET
Structural dynamics of proteins is crucial for understanding the mechanism of the protein functions. Subdomain motions in protein often happen at sub-millisecond, which is hard to be resolved clearly by conventional single molecule FRET experiments. One way to overcome the limit is to take the advantage of the interval photon time measured at nanosecond precision and utilize stochastic data analysis methods, such as maximum likelihood estimation.
We are applying this method to study the structural dynamics of G-protein coupled receptor (GPCR). GPCRs is a large membrane protein family regulate various signal transductions throughout the human body. GPCRs are the targets of 35% of current approved drugs and a large portion of drugs in development. However, the details structural dynamics of the GPCRs upon the binding of ligands are largely unresolved. The difference in the structural dynamics is the key to understanding source of the drug efficacy and bias trigged with different ligands.